Fluorescence in situ Hybridization (FISH)
In situ hybridization (ISH) is a powerful technique designed to detect presence or absence, location, integrity and amount of DNA or RNA sequences in tissues, cells or on chromosomes. ISH is based on the detection of specific sequences by pairing of bases (hybridization) on complementary single strands of nucleic acid. Here, one of the strands is a labeled sequence fragment (probe) that binds only to those parts of the genome with highly or completely complementary sequence, and the other strand is present in the sample material that is to be analyzed. Accordingly, in situ hybridization starts with preparing the sample to be analyzed and with preparing the probe. The typically double-stranded DNA in the sample has to be melted (denatured) into single strands, and the probe has to be labeled to enable detection.
The basic steps for ISH were developed in the 1960s using radioactive isotopes for probe detection. Starting in the early 1980s, protocols using fluorescent dyes for probe visualization have been developed. Because these labels are safer, more stable and easier to handle, the vast majority of in situ hybridization today is done using fluorescence ISH (FISH) protocols. One other important advantage of FISH is that it allows – by using several different fluorescent dyes – the simultaneous detection of different target structures within a single sample.
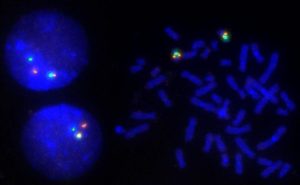
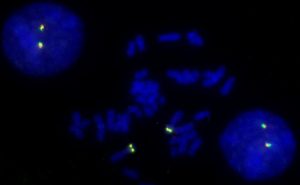
FISH has become a central cytogenetic tool. Probe fragments can be derived from various sources, by recombinant methods or by PCR. Today, accurately mapped bacterial artificial chromosomes (BAC) are the most frequently used probe type. Designing or selecting the right probe for FISH often requires a balance between sensitivity and specificity, i.e. a probe must be large enough to assure specific binding and to carry a sufficient amount of label for detection. On the other hand, with increasing length of the probe the possibility of hybridization to additional target sequences increases as well. In addition, if a probe is too long it may result in a large or diffuse signal.
Probe labeling can be done by linking fluorescent molecules directly to the probe or by tagging probe with antigenic or chemical linkers that will later bind to fluorophores. Depending on which protocol is used, the process can take anywhere from a few hours to a few days. It is called “in situ” to stress the fact that detection of target nucleic acid occurs not on isolated molecules in test tubes, but directly on fresh or fixed cells or tissues.
With FISH, chromosomal rearrangements or aberrations of as small as a few megabase pairs (million base pairs) can be made visible on metaphase (condensed) chromosomes. The resolution goes down to about 1Mb for interphase (spread out) chromosomes. Even greater resolution can be achieved with recently developed methods encompassing artificial stretching of chromatin fibers.
FISH methods cannot detect base-level mutations, e.g. the point mutations underlying some heritable disorders. However, FISH techniques have made landmark contributions in moving the genetic and physical maps of the human genome closer together.
In research, FISH is used to identify the position of genes, determine the number and integrity of human chromosomes, for karyotyping and chromosome painting. In the clinical laboratory, FISH has become a standard tool for detecting chromosomal aberrations, particularly in prenatal and cancer diagnosis. FISH probes and protocols for many genes have been analytically and clinically validated, and many more are being developed and tested.
